Table of Contents
Tests used to identify Gram Positive Bacteria
Mannitol Salt Agar (MSA)

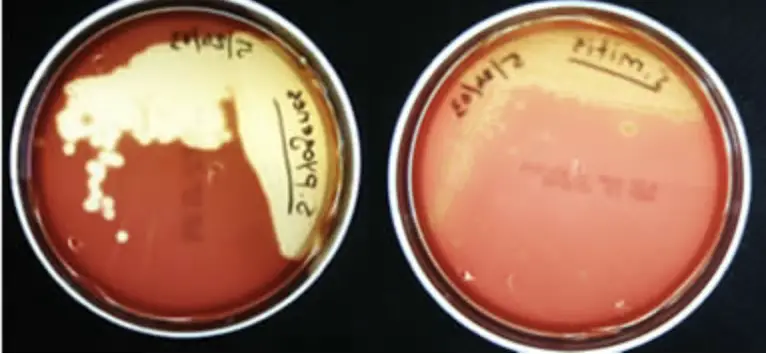
This type of medium is both selective and differential. The MSA will select for organisms such as Staphylococcus species which can live in areas of high salt concentration (plate on the left in the picture below). This is in contrast to Streptococcus species, whose growth is selected against by this high salt agar (plate on the right in the picture below).
The differential ingredient in MSA is the sugar mannitol. Organisms capable of using mannitol as a food source will produce acidic byproducts of fermentation that will lower the pH of the media. The acidity of the media will cause the pH indicator, phenol red, to turn yellow. Staphylococcus aureus is capable of fermenting mannitol (left side of left plate) while Staphylococcus epidermidis is not (right side of left plate).
Glucose broth with Durham tubes
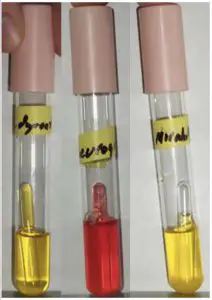 This is a differential medium. It tests an organism’s ability to ferment the sugar glucose as well as its ability to convert the end product of glycolysis, pyruvic acid into gaseous byproducts. This is a test commonly used when trying to identify Gram-negative enteric bacteria, all of which are glucose fermenters but only some of which produce gas.
This is a differential medium. It tests an organism’s ability to ferment the sugar glucose as well as its ability to convert the end product of glycolysis, pyruvic acid into gaseous byproducts. This is a test commonly used when trying to identify Gram-negative enteric bacteria, all of which are glucose fermenters but only some of which produce gas.
Like MSA, this medium also contains the pH indicator, phenol red. If an organism is capable of fermenting the sugar glucose, then acidic byproducts are formed and the pH indicator turns yellow. Escherichia coli is capable of fermenting glucose as are Proteus mirabilis (far right) and Shigella dysenteriae (far left). Pseudomonas aeruginosa (center) is a nonfermenter.
The end product of glycolysis is pyruvate. Organisms that are capable of converting pyruvate to formic acid and formic acid to H2 (g) and CO2 (g), via the action of the enzyme formic hydrogen lyase, emit gas. This gas is trapped in the Durham tube and appears as a bubble at the top of the tube. Escherichia coli and Proteus mirabilis (far right) are both gas producers. Notice that Shigella dysenteriae (far left) ferments glucose but does not produce gas.
*Note – broth tubes can be made containing sugars other than glucose (e.g. lactose and mannitol). Because the same pH indicator (phenol red) is also used in these fermentation tubes, the same results are considered positive (e.g. a lactose broth tube that turns yellow after incubation has been inoculated with an organism that can ferment lactose).
Blood Agar Plates (BAP)
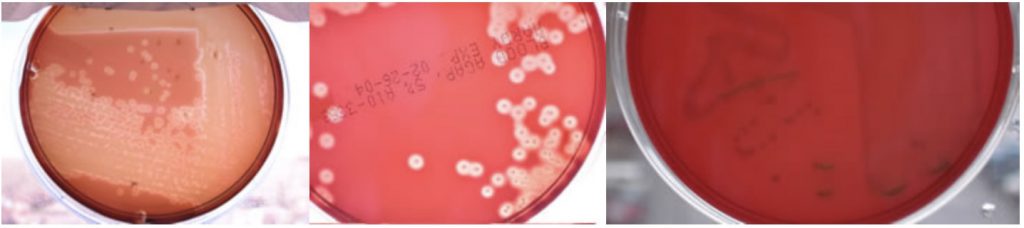
This is a differential medium. It is a rich, complex medium that contains 5% sheep red blood cells. BAP tests the ability of an organism to produce hemolysins, enzymes that damage/lyse red blood cells (erythrocytes). The degree of hemolysis by these hemolysins is helpful in differentiating members of the genera Staphylococcus, Streptococcus and Enterococcus.
- Beta-hemolysis is complete hemolysis. It is characterized by a clear (transparent) zone surrounding the colonies. Staphylococcus aureus, Streptococcus pyogenes and Streptococcus agalactiaeare b-hemolytic (the picture on the left below shows the beta-hemolysis of S. pyogenes).
- Partial hemolysis is termed alpha-hemolysis. Colonies typically are surrounded by a green, opaque zone. Streptococcus pneumoniae and Streptococcus mitis are a-hemolytic (the picture on the right below shows the a-hemolysis of S. mitis).
- If no hemolysis occurs, this is termed gamma-hemolysis. There are no notable zones around the colonies. Staphylococcus epidermidis is gamma-hemolytic.
What type of hemolysis is seen on each one of the following plates?
Streak-stab technique
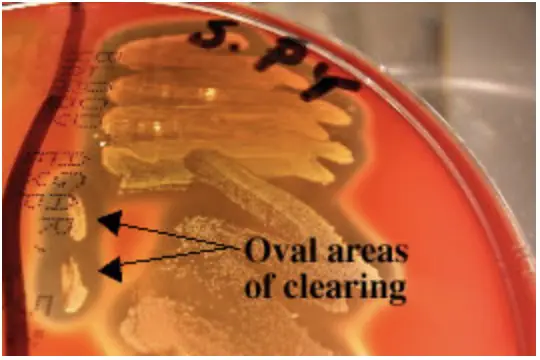 Often when inoculating a BAP to observe hemoloysis patterns, investigators will also stab several times through the agar using an inoculating loop. This stab allows for the detection of streptolysin O, a specific hemolysin produced by Streptococcus pyogenes. This hemolysin is inactivated by O2 and is only seen subsurface (in an anaerobic environment) around the stab mark. Note the oval-shaped areas of clearing around the stab marks in the picture below; these are caused by streptolysin O.
Often when inoculating a BAP to observe hemoloysis patterns, investigators will also stab several times through the agar using an inoculating loop. This stab allows for the detection of streptolysin O, a specific hemolysin produced by Streptococcus pyogenes. This hemolysin is inactivated by O2 and is only seen subsurface (in an anaerobic environment) around the stab mark. Note the oval-shaped areas of clearing around the stab marks in the picture below; these are caused by streptolysin O.
Bile Esculin Agar
 This is a medium that is both selective and differential. It tests the ability of organisms to hydrolyze esculin in the presence of bile. It is commonly used to identify members of the genus Enterococcus (E faecalis and E. faecium).
This is a medium that is both selective and differential. It tests the ability of organisms to hydrolyze esculin in the presence of bile. It is commonly used to identify members of the genus Enterococcus (E faecalis and E. faecium).
The first selective ingredient in this agar is bile, which inhibits the growth of Gram-positives other than enterococci and some streptococci species. The second selective ingredient is sodium azide. This chemical inhibits the growth of Gram-negatives.
The differential ingredient is esculin. If an organism can hydrolyze esculin in the presence of bile, the product esculetin is formed. Esculetin reacts with ferric citrate (in the medium), forming a phenolic iron complex which turns the entire slant dark brown to black. The tube on the far right was inoculated with E. faecalis (positive). The tube in the center was inoculated with a bilie esculin negative organism and the tube on the left was uninoculated.
Sulfur Indole Motility Media (SIM)
 This is a differential medium. It tests the ability of an organism to do several things: reduce sulfur, produce indole and swim through the agar (be motile). SIM is commonly used to differentiate members of Enterobacteriaceae.
This is a differential medium. It tests the ability of an organism to do several things: reduce sulfur, produce indole and swim through the agar (be motile). SIM is commonly used to differentiate members of Enterobacteriaceae.
Sulfur can be reduced to H2S (hydrogen sulfide) either by catabolism of the amino acid cysteine by the enzyme cysteine desulfurase or by reduction of thiosulfate in anaerobic respiration. If hydrogen sulfide is produced, a black color forms in the medium. Proteus mirabilis is positive for H2S production. The organism pictured on the far left is positive for hydrogen sulfide production.
Bacteria that have the enzyme tryptophanase, can convert the amino acid, tryptophane to indole. Indole reacts with added Kovac’s reagent to form rosindole dye which is red in color (indole +). Escherichia coli is indole positive. The organism pictured second from left is E. coli and is indole positive.
SIM tubes are inoculated with a single stab to the bottom of the tube. If an organism is motile than the growth will radiate from the stab mark and make the entire tube appear turbid. Pseudomonas aeruginosa and the strain of Proteus mirabilis that we work with are motile.
Kliger’s Iron Agar (KIA)
 This is a differential medium. It tests for organisms’ abilities to ferment glucose and lactose to acid and acid plus gas end products. It also allows for identification of sulfur reducers. This media is commonly used to separate lactose fermenting members of the family Enterobacteriaceae (e.g. Escherichia coli) from members that do not ferment lactose, like Shigella dysenteriae. These lactose nonfermenting enterics generally tend to be the more serious pathogens of the the gastrointestinal tract.
This is a differential medium. It tests for organisms’ abilities to ferment glucose and lactose to acid and acid plus gas end products. It also allows for identification of sulfur reducers. This media is commonly used to separate lactose fermenting members of the family Enterobacteriaceae (e.g. Escherichia coli) from members that do not ferment lactose, like Shigella dysenteriae. These lactose nonfermenting enterics generally tend to be the more serious pathogens of the the gastrointestinal tract.
The first differential ingredient, glucose, is in very short supply. Organisms capable of fermenting this sugar will use it up within the first few hours of incubation. Glucose fermentation will create acidic byproducts that will turn the phenol red indicator in the media yelllow. Thus, after the first few hours of incubation, the tube will be entirely yellow. At this point, when the glucose has been all used up, the organism must choose another food source. If the organism can ferment lactose, this is the sugar it will choose. Lactose fermentation will continue to produce acidic byproducts and the media will remain yellow (picture on the far left below). If gas is produced as a result of glucose or lactose fermentation, then fissures will appear in the agar or the agar will be lifted off the bottom of the tube.
If an organism cannot use lactose as a food source it will be forced to use the amino acids / proteins in the media. The deamination of the amino acids creates NH3, a weak base, which causes the medium to become alkaline. The alkaline pH causes the phenol red indicator to begin to turn red. Since the incubation time is short (18-24 h), only the slant has a chance to turn red and not the entire tube. Thus an organism that can ferment glucose but not lactose, will produce a red slant and a yellow butt in a KIA tube (second from the left below). These organisms are the more serious pathogens of the GIT such as Shigella dysenteriae.
If an organism is capable of using neither glucose nor lactose, the organism will use solely amino acids / proteins. The slant of the tube will be red and the color of the butt will remain unchanged (picture on the far right below). Pseudomonas aeruginosa is an example of a nonfermenter.
KIA tubes are also capable of detecting the production of H2S. It is seen as a black precipitate (second picture from the right). Sometimes the black precipitate obscures the butt of the tube. In such cases, the organisms should be considered positive for glucose fermentation (yellow butt). Proteus mirabilis (pictured here, second from right) is a glucose positive, lactose negative, sulfur reducing enteric.
Nitrate Broth
This is a differential medium. It is used to determine if an organism is capable of reducing nitrate (NO3-) to nitrite (NO2-) or other nitrogenous compounds via the action of the enzyme nitratase (also called nitrate reductase). This test is important in the identification of both Gram-positive and Gram-negative species.
After incubation, these tubes are first inspected for the presence of gas in the Durham tube. In the case of nonfermenters, this is indicative of reduction of nitrate to nitrogen gas. However, in many cases gas is produced by fermentation and further testing is necessary to determine if reduction of nitrate has occurred. This further testing includes the addition of sulfanilic acid (often called nitrate I) and dimethyl-alpha-napthalamine (nitrate II). If nitrite is present in the media, then it will react with nitrate I and nitrate II to form a red compound. This is considered a positive result.
If no red color forms upon addition of nitrate I and II, this indicates that either the NO3- has not been converted to NO2- (a negative result), or that NO3- was converted to NO2- and then immediately reduced to some other, undetectable form of nitrogen (also a positive result). In order to determine which of the preceding is the case, elemental zinc is added to the broth. Zinc will convert any remaining NO3- to NO2- thus allowing nitrate I and nitrate II to react with the NO2- and form the red pigment (a verified negative result). If no color change occurs upon addition of zinc then this means that the NO3- was converted to NO2- and then was converted to some other undetectable form of nitrogen (a positive result).
If the nitrate broth turns red (tubes pictured in the center) after nitrate I and nitrate II are added, this color indicates a positive result. If instead, the tube turns red (tube pictured on the left) after the addition of Zn, this indicates a negative result. If there is no color change in the tube after the addition of nitrate I and nitrate II, the result is uncertain. If the tube is colorless (picture on the right) after the addition of Zn this indicates a positive test.
Catalase Test
This test is used to identify organisms that produce the enzyme, catalase. This enzyme detoxifies hydrogen peroxide by breaking it down into water and oxygen gas.
The bubbles resulting from production of oxygen gas clearly indicate a catalase positive result. The sample on the right below is catalase positive. The Staphylococcus spp. and the Micrococcus spp. arecatalase positive. The Streptococcus and Enterococcus spp. are catalase negative.
Tests used to identify Gram Negative Bacteria
Oxidase Test
This test is used to identify microorganisms containing the enzyme cytochrome oxidase (important in the electron transport chain). It is commonly used to distinguish between oxidase negative Enterobacteriaceae and oxidase positive Pseudomadaceae.
Cytochrome oxidase transfers electrons from the electron transport chain to oxygen (the final electron acceptor) and reduces it to water. In the oxidase test, artificial electron donors and acceptors are provided. When the electron donor is oxidized by cytochrome oxidase it turns a dark purple. This is considered a positive result. In the picture below the organism on the right (Pseudomonas aeruginosa) is oxidase positive.
Coagulase test
Coagulase is an enzyme that clots blood plasma. This test is performed on Gram-positive, catalase positive species to identify the coagulase positive Staphylococcus aureus. Coagulase is a virulence factor of S. aureus. The formation of clot around an infection caused by this bacteria likely protects it from phagocytosis. This test differentiates Staphylococcus aureus from other coagulase negative Staphylococcus species.
Taxos A (bacitracin sensitivity testing)
This is a differential test used to distinguish between organisms sensitive to the antibiotic bacitracin and those not. Bacitracin is a peptide antibiotic produced by Bacillus subtilis. It inhibits cell wall synthesis and disrupts the cell membrane. This test is commonly used to distinguish between the b-hemolytic streptococci: Streptococcus agalactiae (bacitracin resistant) and Streptococcus pyogenes(bacitracin sensitive). The plate below was streaked with Streptococcus pyogenes; notice the large zone of inhibition surrounding the disk.
Taxos P (optochin sensitivity testing)
This is a differential test used to distinguish between organisms sensitive to the antibiotic optochin and those not. This test is used to distinguish Streptococcus pneumoniae (optochin sensitive (pictured on the right below)) from other a-hemolytic streptococci (optochin resistant (Streptococcus mitis is pictured on the left below)).
MacConkey agar
This medium is both selective and differential. The selective ingredients are the bile salts and the dye, crystal violet which inhibit the growth of Gram-positive bacteria. The differential ingredient is lactose. Fermentation of this sugar results in an acidic pH and causes the pH indicator, neutral red, to turn a bright pinky-red color. Thus organisms capable of lactose fermentation such as Escherichia coli, form bright pinky-red colonies (plate pictured on the left here). MacConkey agar is commonly used to differentiate between the Enterobacteriaceae.
Organism on left is positive for lactose fermentation and that on the right is negative.
Simmon’s Citrate Agar
This is a defined medium used to determine if an organism can use citrate as its sole carbon source. It is often used to differentiate between members of Enterobacteriaceae. In organisms capable of utilizing citrate as a carbon source, the enzyme citrase hydrolyzes citrate into oxaoloacetic acid and acetic acid. The oxaloacetic acid is then hydrolyzed into pyruvic acid and CO2. If CO2 is produced, it reacts with components of the medium to produce an alkaline compound (e.g. Na2CO3). The alkaline pH turns the pH indicator (bromthymol blue) from green to blue. This is a positive result (the tube on the right is citrate positive). Klebsiella pneumoniae and Proteus mirabilis are examples of citrate positive organisms. Escherichia coli and Shigella dysenteriae are citrate negative.
Spirit Blue agar
This agar is used to identify organisms that are capable of producing the enzyme lipase. This enzyme is secreted and hydrolyzes triglycerides to glycerol and three long chain fatty acids. These compounds are small enough to pass through the bacterial cell wall. Glycerol can be converted into a glycolysis intermediate. The fatty acids can be catabolized and their fragments can eventually enter the Kreb’s cycle. Spirit blue agar contains an emulsion of olive oil and spirit blue dye. Bacteria that produce lipase will hydrolyze the olive oil and produce a halo around the bacterial growth. The Gram-positive rod, Bacillus subtilis is lipase positive (pictured on the right) The plate pictured on the left is lipase negative.
Starch hydrolysis test
This test is used to identify bacteria that can hydrolyze starch (amylose and amylopectin) using the enzymes a-amylase and oligo-1,6-glucosidase. Often used to differentiate species from the genera Clostridium and Bacillus. Because of the large size of amylose and amylopectin molecules, these organisms can not pass through the bacterial cell wall. In order to use these starches as a carbon source, bacteria must secrete a-amylase and oligo-1,6-glucosidase into the extracellular space. These enzymes break the starch molecules into smaller glucose subunits which can then enter directly into the glycolytic pathway. In order to interpret the results of the starch hydrolysis test, iodine must be added to the agar. The iodine reacts with the starch to form a dark brown color. Thus, hydrolysis of the starch will create a clear zone around the bacterial growth. Bacillus subtilis is positive for starch hydrolysis (pictured below on the left). The organism shown on the right is negative for starch hydrolysis.
Methyl Red / Voges-Proskauer (MR/VP)
This test is used to determine which fermentation pathway is used to utilize glucose. In the mixed acid fermentation pathway, glucose is fermented and produces several organic acids (lactic, acetic, succinic, and formic acids). The stable production of enough acid to overcome the phosphate buffer will result in a pH of below 4.4. If the pH indicator (methyl red) is added to an aliquot of the culture broth and the pH is below 4.4, a red color will appear (first picture, tube on the left).
If the MR turns yellow, the pH is above 6.0 and the mixed acid fermentation pathway has not been utilized (first picture, tube on the right). The 2,3 butanediol fermentation pathway will ferment glucose and produce a 2,3 butanediol end product instead of organic acids. In order to test this pathway, an aliquot of the MR/VP culture is removed and a-naphthol and KOH are added. They are shaken together vigorously and set aside for about one hour until the results can be read.
The Voges-Proskauer test detects the presence of acetoin, a precursor of 2,3 butanediol. If the culture is positive for acetoin, it will turn “brownish-red to pink” (tube on the left in the second picture). If the culture is negative for acetoin, it will turn “brownish-green to yellow” (tube on the left in the second picture). Note: A culture will usually only be positive for one pathway: either MR+ or VP+. Escherichia coli is MR+ and VP-. In contrast, Enterobacter aerogenes and Klebsiella pneumoniae are MR- and VP+. Pseudomonas aeruginosa is a glucose nonfermenter and is thus MR- and VP-.
CAMP Test
CAMP factor is a diffusible, heat-stable protein produced by group B streptococci. This is a synergistic test between Staphylococcus aureus and Streptococcus agalactiae. S. agalactiae produces CAMP factor. S. aureus produces sphingomyelin C, which binds to red blood cell membranes. The two bacteria are streaked at 90o angles of one another. They do NOT touch. The CAMP factor produced by S. agalactiae enhances the beta-hemolysis of S. aureus by binding to already damaged red blood cells. As a result, an arrow of beta-hemolysis is produced between the two streaks. The test is presumptive for S. agalactiae that produces CAMP factor.
In the picture here, Streptococcus agalactiae was streaked throughout the top region of the plate and brought down toward the center of the plate. Staphylococcus aureus was streaked in a straight line across the center of the plate. Rings of hemolysis are evident all around S. aureus, however the hemolysis if greatly enhanced (in an arrow shape) where the S. agalactiae crosses the hemolysis rings.
Urease test
This test is used to identify bacteria capable of hydrolyzing urea using the enzyme urease. It is commonly used to distinguish the genus Proteus from other enteric bacteria. The hydrolysis of urea forms the weak base, ammonia, as one of its products. This weak base raises the pH of the media above 8.4 and the pH indicator, phenol red, turns from yellow to pink. Proteus mirabilis is a rapid hydrolyzer of urea (center tube pictured here). The tube on the far right was inoculated with a urease negative organism and the tube on the far left was uninoculated.
Motility agar
is a differential medium used to determine whether an organism is equipped with flagella and thus capable of swimming away from a stab mark. The results of motility agar are often difficult to interpret. Generally, if the entire tube is turbid, this indicates that the bacteria have moved away from the stab mark (are motile). The organisms in the two tubes pictured on the right are motile. If, however, the stab mark is clearly visible and the rest of the tube is not turbid, the organism is likely nonmotile (tube pictured on the left).